Section: New Results
Computational Anatomy
Longitudinal brain morphometry: statistical analysis and robust quantification of anatomical changes
Participants : Marco Lorenzi [Correspondant] , Xavier Pennec, Nicholas Ayache.
Longitudinal analysis, Alzheimer's Disease, non-linear registration, brain morphometry
This project is based on the PhD thesis defended in 2012 by Marco Lorenzi, and aims at developing robust and effective instruments for the analysis of longitudinal brain changes, with special focus on the study of brain atrophy in Alzheimer's disease. The project relies on the analysis of follow-up magnetic resonance images of the brain by means of non-linear registration. During 2013 the main scientific achievements were the following:
-
We developed and distributed the LCC-logDemons, an accurate and robust diffeomorphic non-linear registration algorithm [14] , [16] . The algorithm implements the symmetric Local Correlation Coefficient (LCC) and is suited for both inter and intra-subject registration. The software is freely available for research purposes here .
-
We investigated the problem of comparing the trajectories of longitudinal morphological changes estimated in different patients. Based on our previous work on parallel transport in diffeomorphic registration, we proposed the "pole ladder" for the efficient normalization of longitudinal trajectories in a common reference space [15] , [48] .
-
We defined an effective framework for the statistical analysis of longitudinal brain changes in clinical groups. The proposed framework enabled the characterization of abnormal morphological changes in healthy subjects at risk for Alzheiemer's disease [46] .
-
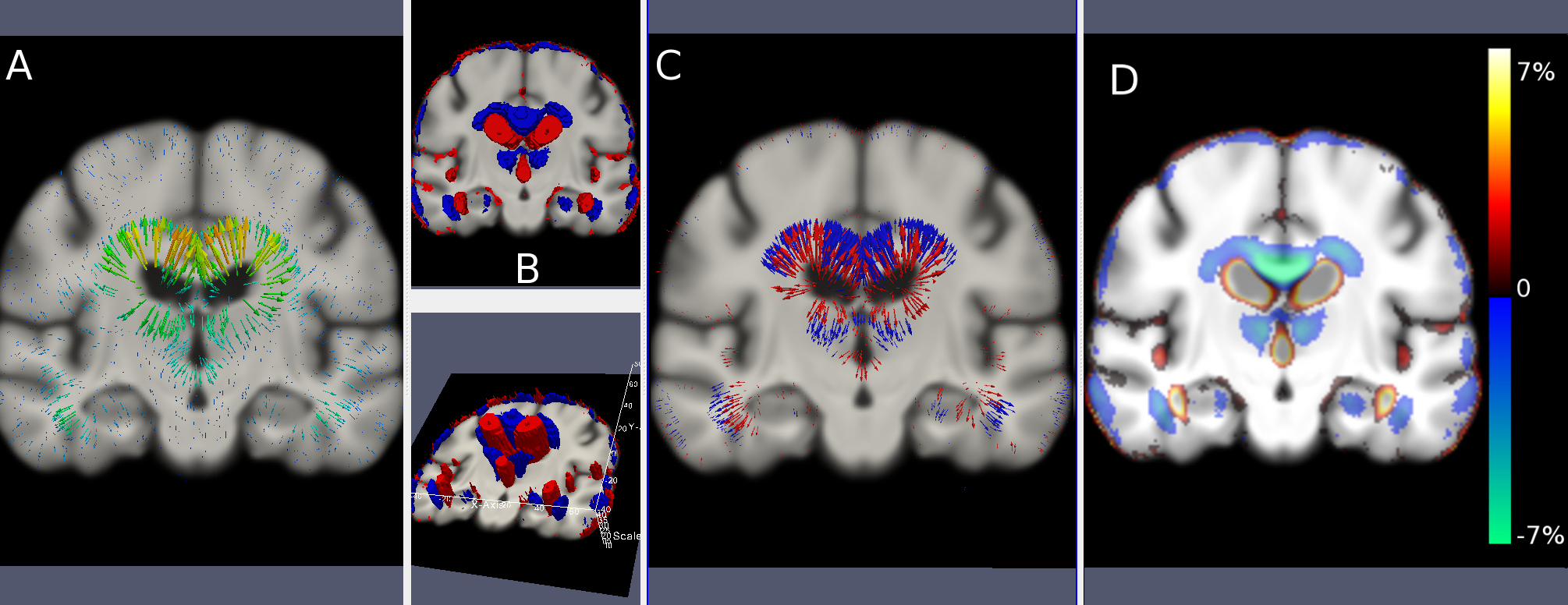
We addressed the multi-scale analysis of longitudinal volume changes encoded by deformation fields. We provided a probabilistic framework for the consistent definition of anatomical regions of longitudinal brain atrophy across spatial scales, in order to robustly quantify regional volume changes in populations or in single patients. The framework was applied to the longitudinal analysis of group-wise atrophy in Alzheimer's disease (Figure 5 ), and to the tracking and quantification of treatment efficacy on brain tumors [47] .
Longitudinal Analysis and Modeling of Brain Development during Adolescence
Participants : Mehdi Hadj-Hamou [Correspondant] , Xavier Pennec, Nicholas Ayache.
This work is partly funded through the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Brain development, adolescence, longitudinal analysis, non-rigid registration algorithm
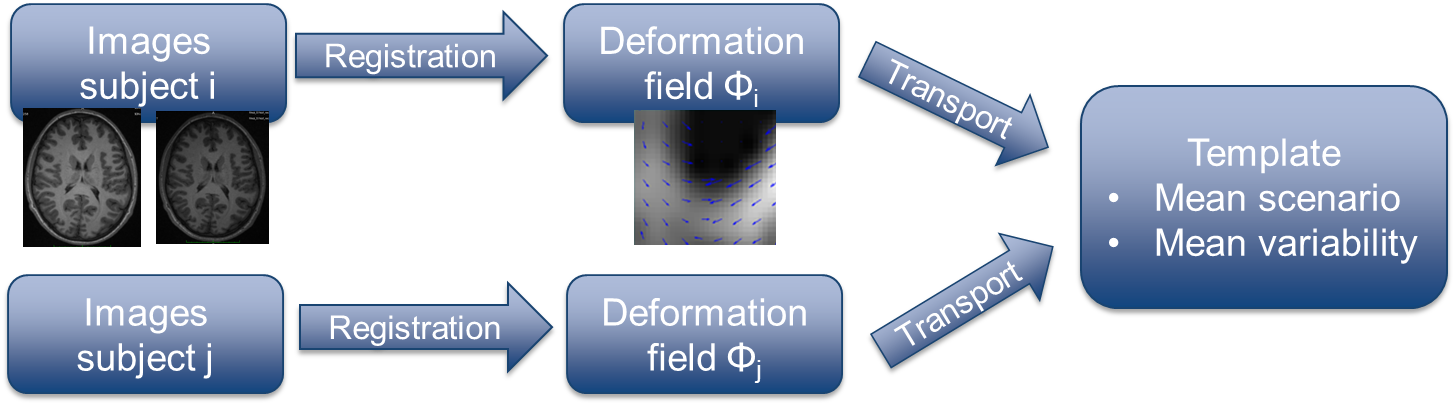
Due to the lack of tools to capture the subtle changes in the brain, little is known about its development during adolescence. The aim of this project is then to provide quantification and models of brain development during adolescence based on non-rigid registration of longitudinal MRIs (enabling us to capture these changes). The analysis pipeline is the following (Figure 6 ) :
-
Register each patient's pair of images in order to get access to the longitudinal changes defined by a transformation field (parameterized by a Stationary Velocity Field).
-
Transport every deformation field in a common space (template) to obtain the mean scenario and quantify the changes.
-
Propose simplified models of the anatomical changes occurring during adolescence abstracting the results of the analysis.
Reduced-Order Statistical Models of Cardiac Growth, Motion and Blood Flow
Participants : Kristin Mcleod [Correspondant] , Maxime Sermesant, Xavier Pennec.
This work was partially funded by the EU projects Care4me (ITEA2) and MD-Paedigree (FP7).
Statistical analysis, image registration, Demons algorithm, reduced models, CFD, Polyaffine, cardiac motion tracking
This work involves developing reduced models of cardiac growth, motion and blood flow, with application to the Tetralogy of Fallot heart [28] .
-
Extending the 2012 reduced order model of cardiac motion based on a polyaffine log-demons registration proposed at the 2012 STACOM MICCAI workshop, an additional cardiac-specific prior was added to the model to give more physiologically meaningful weight functions. Using this method, the trace of the affine matrix per region was plotted over time to establish differences between healthy subjects and asynchronous heart failure patients. The method and results were presented at the 2013 FIMH conference [52] .
-
Going further in analysing the affine parameters per region, statistical methods were applied to the registration parameters of the method proposed at the 2012 STACOM MICCAI workshop [50] . By applying principal component analysis to the transformation parameters stacked either column-wise or row-wise, population-based descriptors of motion in terms of the temporal or spatial components were obtained. The method was applied to 15 healthy subjects and 2 heart failure patients and presented at the 2013 MICCAI conference [51] .
-
The analysis of a statistical model for reduced blood flow simulations in the pulmonary artery proposed in the 2010 STACOM workshop was extended to a journal version [10] , [64] . The previous work was extended to re-solve the obtained pressure and velocity bases for the subject-specific geometry by solving the Navier Stokes equations on the reduced bases. The method was applied to a data-set of 17 Tetralogy of Fallot patients.
Geometric Statistics
Participants : Xavier Pennec [Correspondant] , Nina Miolane, Christof Seiler [Stanford] , Susan Holmes [Stanford] .
This work is partly funded through a France Stanford collaborative project grant (2013-2014).
Statistics, manifolds, Lie groups
The study of bi-invariant means on Lie groups [53] was further pushed by looking for the conditions of existence of bi-invariant semi-Riemannian metrics, thus relaxing the positivity constraint of Riemannian metrics [4] . This idea was based on the fact that such a bi-invariant semi-Riemannian metric exists of SE(3). Unfortunately, this does not generalize to higher dimensions. Other results on geometric statistics on regions for in the context of group-valued trees for deformation analysis were presented in [55] .